Epidemiologists predicting the spread of Covid-19 should adopt climate-modelling methods to make forecasts more reliable, say computer scientists who have spent months auditing one of the most influential models of the pandemic.
In a study that was uploaded to the preprint platform Research Square on November 6, researchers commissioned by London’s Royal Society used a powerful supercomputer to re-examine CovidSim, a model developed by a group at Imperial College London.
In March, that simulation helped convince British and US politicians to introduce lockdowns to prevent projected deaths, but it has since been scrutinised by researchers who doubt the reliability of its results.
The analysis, which has not yet been peer-reviewed, shows that because researchers did not appreciate how sensitive CovidSim was to small changes in its inputs, their results overestimated the extent to which a lockdown was likely to reduce deaths, says Peter Coveney, a chemist and computer scientist at University College London, who led the study.
Coveney is reluctant to criticise the Imperial group, led by epidemiologist Neil Ferguson, which he says did the best job possible under the circumstances. And the model correctly showed that “doing nothing at all would have disastrous consequences”, he says. But he argues that epidemiologists should stress-test their simulations by running ‘ensemble’ models, in which thousands of versions of the model are run with a range of assumptions and inputs, to provide a spread of scenarios with different probabilities.
These ‘probabilistic’ methods are routine in computation-heavy fields, from weather forecasting to molecular dynamics. Coveney’s team has now done this for CovidSim. The findings suggest that if the model had been run as an ensemble, it would have forecast a range of probable death tolls under lockdown, with an average twice as high as the original prediction, and closer to the actual figures.
“CovidSim may be vaunted as the most complicated epidemiological model, but it is almost like a toy compared with the really high-end supercomputing applications,” says Coveney, who was asked to check the model’s performance as part of the Royal Society’s Rapid Assistance in Modelling the Pandemic (RAMP) initiative.
Coveney’s team used the Eagle supercomputer at the Poznan Supercomputing and Networking Centre in Poland to perform 6,000 separate runs of CovidSim, each with a unique set of input parameters. These represent features of the pandemic including the infectiousness and lethality of the virus, the probable number of contacts people make in various settings and the estimated success of measures such as telling people to work from home.
Back in March, inputs for many of these parameters were educated guesses, with some drawn from preliminary data on the virus and others based on experience with diseases such as influenza. Models that predict the spread of disease often rely on hundreds of parameters — but this can introduce uncertainty.
“There was a concern among the circles who set up the RAMP initiative that these models the epidemiologists work with have an absurd number of parameters in them and they can’t possibly be right,” Coveney says.
His team found 940 parameters in the CovidSim code, but whittled these down to the 19 that most affected the output. And up to two-thirds of the differences in the model’s results could be put down to changes in just three key variables: the length of the latent period during which an infected person has no symptoms and can’t pass the virus on; the effectiveness of social distancing; and how long after getting infected a person goes into isolation.
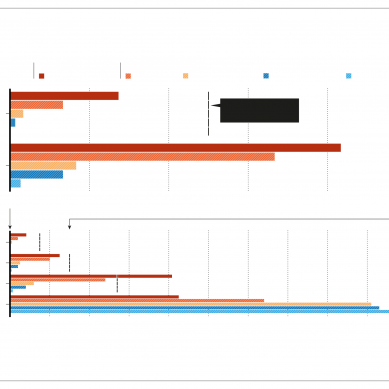
The study suggests that small variations in these parameters could have an outsized, non-linear impact on the model’s output. For example, the majority of the team’s thousands of runs suggested that the UK death toll under lockdown would be much higher than the Imperial team’s initial projections — 5–6 times higher in some cases. Averaging the figures still suggested twice as many deaths as the Imperial group had forecast.
In one modelled scenario, which assumed that the United Kingdom would lock down when 60 people per week needed to be admitted to hospital for intensive care, the March report forecast a total of 8,700 deaths in the country.
The probabilistic results produced by Coveney’s group put this figure at around 15,000 on average, but said that death tolls of more than 40,000 were possible, depending on what parameters were used. It is hard to compare these projections with the actual figures for Covid-19 deaths in the United Kingdom, because the lockdown started a week later than the results of any of the models assume, by which time significantly higher amounts of the disease were already circulating.
“They didn’t get it right,” says Coveney. “They ran the simulation correctly: it’s just that they didn’t know how to extract the correct probabilistic description from it. That would mean having to run ensembles of calculations.”
Coveney said he couldn’t comment on whether running an ensemble model would have altered policy, but Rowland Kao, an epidemiologist and data scientist at the University of Edinburgh, UK, points out that the government compares and synthesises the results of several different Covid-19 models. “It would be overly simplified to consider that decision-making is based on a single model,” he says.
Ferguson accepts most of Coveney’s points about the benefits of performing probabilistic forecasts, but says that “we just weren’t in a position to do that in March”. The Imperial group has significantly improved its models since then, he adds, and can now produce probabilistic results. For example, it now presents the uncertainty in CovidSim inputs using Bayesian statistical tools — already common in some epidemiological models of illnesses such as the livestock disease foot-and-mouth.
And a simpler model, he adds, was used to inform the UK government’s decision to reintroduce lockdown measures in England this month. This model is more agile than CovidSim, “Because we can run it several times a week. It’s much easier to fit the data in real time, allowing for uncertainty,” Ferguson says.
“This sounds like a step in the right direction and is aligned with the conclusions of our paper,” says Coveney.
The choice of technique often comes down to a computational trade-off, Ferguson says. “If you want to routinely properly characterize all the uncertainty, then that is much easier with a less computationally intensive model.”
Bayesian tools are an improvement, says Tim Palmer, a climate physicist at the University of Oxford, UK, who pioneered the use of ensemble modelling in weather forecasting. But only ensemble modelling techniques that are run on the most powerful computers will deliver the most reliable pandemic projections, he says.
Such techniques transformed the reliability of climate models, he adds, helped by the coordination of the Intergovernmental Panel on Climate Change (IPCC).
“We need something like the IPCC for these pandemic models. We need some kind of international facilities where these models can be developed properly,” Palmer says.
He adds, “It has been rushed because of the urgency of the situation. But to take this forward, we need some kind of international organisation that can work on synthesising epidemiological models from around the world.”
- A Nature magazine report